Agilent RNA Gels - Samples 61 through 69, 48 and 49
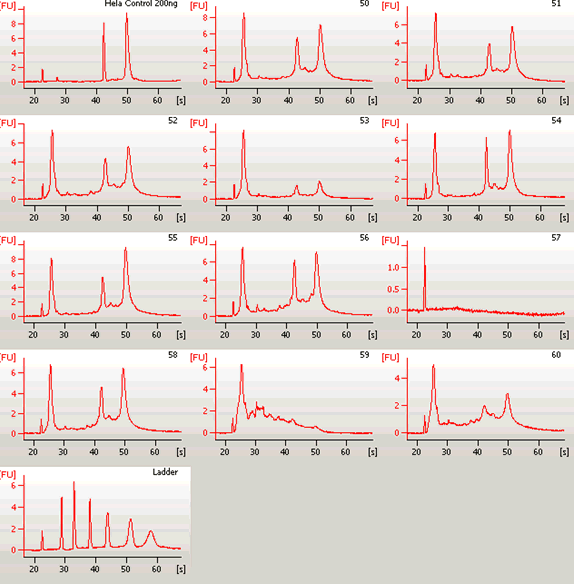
Samples 50 through 60 in consecutive order on Agilent RNA gel
Note: 57 was repeated (see previous page) based on preliminary results from the Gelman laboratory, and satisfactory results were obtained on the repeat.
IDENTITY OFSAMPLES 50 - 60 INCLUDING ORIGINAL #57
BG-A2-50
BG-A3-51
BG-A4-52
BG-A5-53
BG-A6-54
BG-B1-55
BG-B2-56
BG-B3-57
BG-B4-58
BG-B5-59** ¶
BG-B6-60** ¶
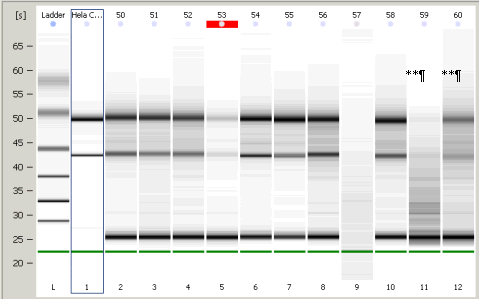
** Denotes that the isolated RNA contained degradation that has the potential to adversely affect reliability of the results of the array. After the results of the hybridization signals were screened for evidence that signal patterns were adversely affected, some degraded specimens were affected adversely ( ¶ ), and some were not.(**). #57 shows evidence of a technical problem and it was repeated on another gel. It produced satisfactory results on the repeat. See above.
¶ Denotes that the hybridization signal pattern of this sample was influenced by RNA degradation when it was analyzed. One or more outcomes were used to screen for anomalous hybridization on a particular chip. See 6.0 for details. Briefly, a hybridization signal was present in a relatively high percentage of all the probe sets, or signal intensities were weak across the board and required a high scaling factor for calibration. Both of those measures generally indicate that probe sets that did not produce a significant hybridization signal in most of the samples as a whole may erroneously report a signal being present on these particular chips. As well spiked control RNA may have shown an uneven distribution of cDNA representation due to uneven conversion to cDNA. This means that the effectiveness of hybridization of some transcripts on the chip, but not all transcripts on the chip, may have been corrupted due to RNA degradation. These errors can produce over- or under-representation of certain signal intensities relative to the other transcripts on the array.
** ¶ Both of the above
BOLD denotes that this case was added to compensate for cases that had preliminary evidence of potentially substantial RNA degradation
Gel traces for samples 50 through 60, including the original trace of #57
#57 was repeated on another Agilent RNA gel and produced a different result.
Agilent RNA gels were run after a column purification step
One sample (HELA) shown in the image of this gel run was not related to this study.
Traces from samples that were adversely affected on the gene array are indicated by including the full NNTC identification number.
Note that #57 shows that a technical malfunction occurred on this run; the sample contained intact RNA on the repeat and it produced reliable hybridization signals.